Stacking layers
Contents
Stacking layers#
Do not see them differently
Do not consider all as the same
Unwaveringly, practice guarding the One
In Geographic Information Systems 🌐, geographic data is arranged as different ‘layers’ 🍰. For example:
Multispectral or hyperspectral 🌈 optical satellites collect different radiometric bands from slices along the electromagnetic spectrum
Synthetic Aperture Radar (SAR) 📡 sensors have different polarizations such as HH, HV, VH & VV
Satellite laser and radar altimeters 🛰️ measure elevation which can be turned into a Digital Elevation Model (DEM)
As long as these layers are georeferenced 📍 though, they can be stacked! This tutorial will cover the following topics:
Searching for spatiotemporal datasets in a dynamic STAC Catalog 📚
Stacking time-series 📆 data into a 4D tensor of shape (time, channel, y, x)
Organizing different geographic 🗺️ layers into a dataset suitable for change detection
🎉 Getting started#
Load up them libraries!
import os
import matplotlib.pyplot as plt
import numpy as np
import planetary_computer
import pystac
import rasterio
import torch
import torchdata
import xarray as xr
import zen3geo
0️⃣ Search for spatiotemporal data 📅#
This time, we’ll be looking at change detection using time-series data. The focus area is Gunung Talamau, Sumatra Barat, Indonesia 🇮🇩 where an earthquake on 25 Feb 2022 triggered a series of landslides ⛰️. Affected areas will be mapped using Sentinel-1 Radiometrically Terrain Corrected (RTC) intensity SAR data 📡 obtained via a spatiotemporal query to a STAC API.
🔗 Links:
This is how the Sentinel-1 radar image looks like over Sumatra Barat, Indonesia on 23 February 2022, two days before the earthquake.

Sentinel-1 PolSAR time-series ⏳#
Before we start, we’ll need to set the PC_SDK_SUBSCRIPTION_KEY environment
variable 🔡 to access the Sentinel-1 RTC data from Planetary Computer 💻. The
steps are:
Get a 🪐 Planetary Computer account at https://planetarycomputer.microsoft.com/account/request
Follow 🧑🏫 instructions to get a subscription key
Go to https://planetarycomputer.developer.azure-api.net/profile. You should have a Primary key 🔑 and Secondary key 🗝️, click on ‘Show’ to reveal it. Copy and paste the key below, or better, set it securely 🔐 in something like a
.envfile!
# Uncomment the line below and set your Planetary Computer subscription key
# os.environ["PC_SDK_SUBSCRIPTION_KEY"] = "abcdefghijklmnopqrstuvwxyz123456"
Done? Let’s now define an 🧭 area of interest and 📆 time range covering one month before and one month after the earthquake ⚠️.
# Spatiotemporal query on STAC catalog for Sentinel-1 RTC data
query = dict(
bbox=[99.8, -0.24, 100.07, 0.15], # West, South, East, North
datetime=["2022-01-25T00:00:00Z", "2022-03-25T23:59:59Z"],
collections=["sentinel-1-rtc"],
)
dp = torchdata.datapipes.iter.IterableWrapper(iterable=[query])
Then, search over a dynamic STAC Catalog 📚 for items matching the
spatiotemporal query ❔ using
zen3geo.datapipes.PySTACAPISearcher (functional name:
search_for_pystac_item).
dp_pystac_client = dp.search_for_pystac_item(
catalog_url="https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)
Tip
Confused about which parameters go where 😕? Here’s some clarification:
Different spatiotemporal queries (e.g. for multiple geographical areas) should go in
torchdata.datapipes.iter.IterableWrapper, e.g.IterableWrapper(iterable=[query_area1, query_area2]). The query dictionaries will be passed topystac_client.Client.search().Common parameters to interact with the STAC API Client should go in
search_for_pystac_item(), e.g. the STAC API’s URL (see https://stacindex.org/catalogs?access=public&type=api for a public list) and connection related parameters. These will be passed topystac_client.Client.open().
The output is a pystac_client.ItemSearch 🔎 instance that only
holds the STAC API query information ℹ️ but doesn’t request for data! We’ll
need to order it 🧞 to return something like a
pystac.ItemCollection.
def get_all_items(item_search) -> pystac.ItemCollection:
return item_search.item_collection()
dp_sen1_items = dp_pystac_client.map(fn=get_all_items)
dp_sen1_items
MapperIterDataPipe
Take a peek 🫣 to see if the query does contain STAC items.
it = iter(dp_sen1_items)
item_collection = next(it)
item_collection.items
[<Item id=S1A_IW_GRDH_1SDV_20220320T230514_20220320T230548_042411_050E99_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220320T230449_20220320T230514_042411_050E99_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220319T114141_20220319T114206_042389_050DD7_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220308T230513_20220308T230548_042236_0508AF_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220308T230448_20220308T230513_042236_0508AF_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220307T114141_20220307T114206_042214_0507E8_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220224T230514_20220224T230548_042061_0502B9_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220224T230449_20220224T230514_042061_0502B9_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220223T114141_20220223T114206_042039_0501F9_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220212T230514_20220212T230548_041886_04FCA5_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220212T230449_20220212T230514_041886_04FCA5_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220211T114141_20220211T114206_041864_04FBD9_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220131T230514_20220131T230548_041711_04F691_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220131T230449_20220131T230514_041711_04F691_rtc>,
<Item id=S1A_IW_GRDH_1SDV_20220130T114142_20220130T114207_041689_04F5D0_rtc>]
Copernicus Digital Elevation Model (DEM) ⛰️#
Since landslides 🛝 typically happen on steep slopes, it can be useful to have a 🏔️ topographic layer. Let’s set up a STAC query 🙋 to get the 30m spatial resolution Copernicus DEM.
# Spatiotemporal query on STAC catalog for Copernicus DEM 30m data
query = dict(
bbox=[99.8, -0.24, 100.07, 0.15], # West, South, East, North
collections=["cop-dem-glo-30"],
)
dp_copdem30 = torchdata.datapipes.iter.IterableWrapper(iterable=[query])
Just to be fancy, let’s chain 🔗 together the next two datapipes.
dp_copdem30_items = dp_copdem30.search_for_pystac_item(
catalog_url="https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
).map(fn=get_all_items)
dp_copdem30_items
MapperIterDataPipe
This is one of the four DEM tiles 🀫 that will be returned from the query.
Landslide extent vector polygons 🔶#
Now for the target labels 🏷️. Following Vector segmentation masks,
we’ll first load the digitized landslide polygons from a vector file 📁 using
zen3geo.datapipes.PyogrioReader (functional name:
read_from_pyogrio).
# https://gdal.org/user/virtual_file_systems.html#vsizip-zip-archives
shape_url = "/vsizip/vsicurl/https://unosat-maps.web.cern.ch/ID/LS20220308IDN/LS20220308IDN_SHP.zip/LS20220308IDN_SHP/S2_20220304_LandslideExtent_MountTalakmau.shp"
dp_shapes = torchdata.datapipes.iter.IterableWrapper(iterable=[shape_url])
dp_pyogrio = dp_shapes.read_from_pyogrio()
dp_pyogrio
PyogrioReaderIterDataPipe
Let’s take a look at the geopandas.GeoDataFrame data table
📊 to see the attributes inside.
it = iter(dp_pyogrio)
geodataframe = next(it)
print(geodataframe.bounds)
geodataframe.dropna(axis="columns")
minx miny maxx maxy
0 99.806331 -0.248744 100.065765 0.147054
| SensorDate | EventCode | Area_m2 | Area_ha | Shape_Leng | Shape_Area | SensorID | Confidence | FieldValid | Main_Dmg | StaffID | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2022-04-03 | LS20220308IDN | 6238480.0 | 623.848 | 2.873971 | 0.000507 | Sentinel-2 | To Be Evaluated | Not yet field validated | Landslide | TH | MULTIPOLYGON (((99.80642 -0.24865, 99.80642 -0... |
We’ll show you what the landslide segmentation masks 😷 look like after it’s been rasterized later 😉.
1️⃣ Stack bands, append variables 📚#
There are now three layers 🍰 to handle, two rasters and a vector. This section will show you step by step 📶 instructions to combine them using xarray like so:
Stack the Sentinel-1 🛰️ time-series STAC Items (GeoTIFFs) into an
xarray.DataArray.Combine the Sentinel-1 and Copernicus DEM ⛰️
xarray.DataArraylayers into a singlexarray.Dataset.Using the
xarray.Datasetas a canvas template, rasterize the landslide 🛝 polygon extents, and append the resulting segmentation mask as another data variable 🗃️ in thexarray.Dataset.
Stack multi-channel time-series GeoTIFFs 🗓️#
Each pystac.Item in a pystac.ItemCollection represents
a 🛰️ Sentinel-1 RTC image captured at a particular datetime ⌚. Let’s subset
the data to just the mountain area, and stack 🥞 all the STAC items into a 4D
time-series tensor using zen3geo.datapipes.StackSTACStacker
(functional name: stack_stac_items)!
dp_sen1_stack = dp_sen1_items.stack_stac_items(
assets=["vh", "vv"], # SAR polarizations
epsg=32647, # UTM Zone 47N
resolution=30, # Spatial resolution of 30 metres
bounds_latlon=[99.933681, -0.009951, 100.065765, 0.147054], # W, S, E, N
xy_coords="center", # pixel centroid coords instead of topleft corner
dtype=np.float16, # Use a lightweight data type
)
dp_sen1_stack
StackSTACStackerIterDataPipe
The keyword arguments are 📨 passed to stackstac.stack() behind the
scenes. The important❕parameters to set in this case are:
assets: The STAC item assets 🍱 (typically the ‘band’ names)
epsg: The 🌐 EPSG projection code, best if you know the native projection
resolution: Spatial resolution 📏. The Sentinel-1 RTC is actually at 10m, but we’ll resample to 30m to keep things small 🤏 and match the Copernicus DEM.
The result is a single xarray.DataArray ‘datacube’ 🧊 with
dimensions (time, band, y, x).
it = iter(dp_sen1_stack)
dataarray = next(it)
dataarray
<xarray.DataArray 'stackstac-3b069ee52dbc03b1fd6c981837d6c185' (time: 15,
band: 2,
y: 579, x: 491)>
dask.array<fetch_raster_window, shape=(15, 2, 579, 491), dtype=float16, chunksize=(1, 1, 579, 491), chunktype=numpy.ndarray>
Coordinates: (12/39)
* time (time) datetime64[ns] 2022-01-30T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2022...
* band (band) <U2 'vh' 'vv'
* x (x) float64 6.039e+05 ... 6.186e+05
* y (y) float64 1.624e+04 ... -1.095e+03
sar:resolution_range int64 20
... ...
sar:looks_equivalent_number float64 4.4
sat:absolute_orbit (time) int64 41689 41711 ... 42411
description (band) <U173 'Terrain-corrected ga...
title (band) <U41 'VH: vertical transmit...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32647
Attributes:
spec: RasterSpec(epsg=32647, bounds=(603870, -1110, 618600, 16260)...
crs: epsg:32647
transform: | 30.00, 0.00, 603870.00|\n| 0.00,-30.00, 16260.00|\n| 0.00,...
resolution: 30Append single-band DEM to datacube 🧊#
Time for layer number 2 💕. Let’s read the Copernicus DEM ⛰️ STAC Item into an
xarray.DataArray first, again via
zen3geo.datapipes.StackSTACStacker (functional name:
stack_stac_items). We’ll need to ensure ✔️ that the DEM is reprojected to the
same 🌏 coordinate reference system and 📐 aligned to the same spatial extent
as the Sentinel-1 time-series.
dp_copdem_stack = dp_copdem30_items.stack_stac_items(
assets=["data"],
epsg=32647, # UTM Zone 47N
resolution=30, # Spatial resolution of 30 metres
bounds_latlon=[99.933681, -0.009951, 100.065765, 0.147054], # W, S, E, N
xy_coords="center", # pixel centroid coords instead of topleft corner
dtype=np.float16, # Use a lightweight data type
resampling=rasterio.enums.Resampling.bilinear, # Bilinear resampling
)
dp_copdem_stack
StackSTACStackerIterDataPipe
it = iter(dp_copdem_stack)
dataarray = next(it)
dataarray
<xarray.DataArray 'stackstac-afdc91bccb12ec4ba1376f87b0641b62' (time: 4,
band: 1,
y: 579, x: 491)>
dask.array<fetch_raster_window, shape=(4, 1, 579, 491), dtype=float16, chunksize=(1, 1, 579, 491), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 2021-04-22 2021-04-22 ... 2021-04-22
id (time) <U40 'Copernicus_DSM_COG_10_S01_00_E100_00_DEM' ... 'C...
* band (band) <U4 'data'
* x (x) float64 6.039e+05 6.039e+05 ... 6.186e+05 6.186e+05
* y (y) float64 1.624e+04 1.622e+04 ... -1.065e+03 -1.095e+03
proj:epsg int64 4326
platform <U8 'TanDEM-X'
gsd int64 30
proj:shape object {3600}
epsg int64 32647
Attributes:
spec: RasterSpec(epsg=32647, bounds=(603870, -1110, 618600, 16260)...
crs: epsg:32647
transform: | 30.00, 0.00, 603870.00|\n| 0.00,-30.00, 16260.00|\n| 0.00,...
resolution: 30Why are there 4 ⏳ time layers? Actually, the STAC query had returned four DEM
tiles 🀫, and stackstac.stack() has stacked both of them along a
dimension name ‘time’ (probably better named ‘tile’). Fear not, the tiles can
be joined 🍒 into a single terrain mosaic layer with dimensions (“band”, “y”,
“x”) using zen3geo.datapipes.StackSTACMosaicker (functional name:
mosaic_dataarray).
dp_copdem_mosaic = dp_copdem_stack.mosaic_dataarray(nodata=0)
dp_copdem_mosaic
StackSTACMosaickerIterDataPipe
Great! The two xarray.DataArray objects (Sentinel-1 and Copernicus
DEM mosaic) can now be combined 🪢. First, use
torchdata.datapipes.iter.Zipper (functional name: zip) to put the
two xarray.DataArray objects into a tuple 🎵.
dp_sen1_copdem = dp_sen1_stack.zip(dp_copdem_mosaic)
dp_sen1_copdem
ZipperIterDataPipe
Next, use torchdata.datapipes.iter.Collator (functional name:
collate) to convert 🤸 the tuple of xarray.DataArray objects into
an xarray.Dataset 🧊, similar to what was done in
Object detection boxes.
def sardem_collate_fn(sar_and_dem: tuple) -> xr.Dataset:
"""
Combine a pair of xarray.DataArray (SAR, DEM) inputs into an
xarray.Dataset with data variables named 'vh', 'vv' and 'dem'.
"""
# Turn 2 xr.DataArray objects into 1 xr.Dataset with multiple data vars
sar, dem = sar_and_dem
# Initialize xr.Dataset with VH and VV channels
dataset: xr.Dataset = sar.sel(band="vh").to_dataset(name="vh")
dataset["vv"] = sar.sel(band="vv")
# Add Copernicus DEM mosaic as another layer
dataset["dem"] = dem.squeeze()
return dataset
dp_vhvvdem_dataset = dp_sen1_copdem.collate(collate_fn=sardem_collate_fn)
dp_vhvvdem_dataset
CollatorIterDataPipe
Here’s how the current xarray.Dataset 🧱 is structured. Notice that
VH and VV polarization channels 📡 are now two separate data variables, each
with dimensions (time, y, x). The DEM ⛰️ data is not a time-series, so it has
dimensions (y, x) only. All the ‘band’ dimensions have been removed ❌ and are
now data variables within the xarray.Dataset 😎.
it = iter(dp_vhvvdem_dataset)
dataset = next(it)
dataset
<xarray.Dataset>
Dimensions: (time: 15, x: 491, y: 579)
Coordinates: (12/41)
* time (time) datetime64[ns] 2022-01-30T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2022...
band <U2 'vh'
* x (x) float64 6.039e+05 ... 6.186e+05
* y (y) float64 1.624e+04 ... -1.095e+03
sar:resolution_range int64 20
... ...
description <U173 'Terrain-corrected gamma nau...
title <U41 'VH: vertical transmit, horiz...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32647
gsd int64 30
proj:shape object {3600}
Data variables:
vh (time, y, x) float16 dask.array<chunksize=(1, 579, 491), meta=np.ndarray>
vv (time, y, x) float16 dask.array<chunksize=(1, 579, 491), meta=np.ndarray>
dem (y, x) float16 dask.array<chunksize=(579, 491), meta=np.ndarray>Visualize the DataPipe graph ⛓️ too for good measure.
torchdata.datapipes.utils.to_graph(dp=dp_vhvvdem_dataset)
Rasterize target labels to datacube extent 🏷️#
The landslide polygons 🔶 can now be rasterized and added as another layer to
our datacube 🧊. Following Vector segmentation masks, we’ll first fork
the DataPipe into two branches 🫒 using
torchdata.datapipes.iter.Forker (functional name: fork).
dp_vhvvdem_canvas, dp_vhvvdem_datacube = dp_vhvvdem_dataset.fork(num_instances=2)
dp_vhvvdem_canvas, dp_vhvvdem_datacube
(_ChildDataPipe, _ChildDataPipe)
Next, create a blank canvas 📃 using
zen3geo.datapipes.XarrayCanvas (functional name:
canvas_from_xarray) and rasterize 🖌 the vector polygons onto the template
canvas using zen3geo.datapipes.DatashaderRasterizer (functional
name: rasterize_with_datashader)
dp_datashader = dp_vhvvdem_canvas.canvas_from_xarray().rasterize_with_datashader(
vector_datapipe=dp_pyogrio
)
dp_datashader
DatashaderRasterizerIterDataPipe
Cool, and this layer can be added 🧮 as another data variable in the datacube.
def cubemask_collate_fn(cube_and_mask: tuple) -> xr.Dataset:
"""
Merge target 'mask' (xarray.DataArray) into an existing datacube
(xarray.Dataset) as another data variable.
"""
datacube, mask = cube_and_mask
merged_datacube = xr.merge(objects=[datacube, mask.rename("mask")], join="override")
return merged_datacube
dp_datacube = dp_vhvvdem_datacube.zip(dp_datashader).collate(
collate_fn=cubemask_collate_fn
)
dp_datacube
CollatorIterDataPipe
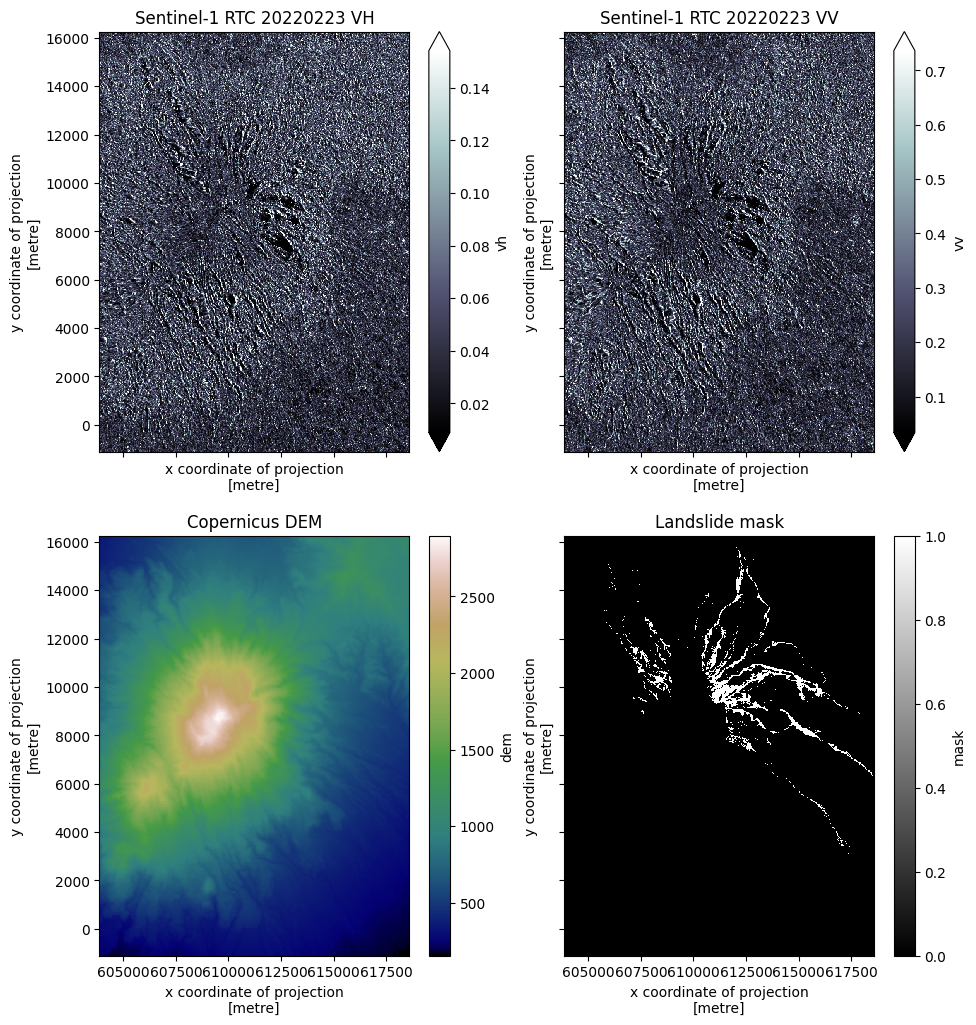
Inspect the datacube 🧊 and visualize all the layers 🧅 within.
it = iter(dp_datacube)
datacube = next(it)
datacube
<xarray.Dataset>
Dimensions: (time: 15, x: 491, y: 579)
Coordinates: (12/42)
* time (time) datetime64[ns] 2022-01-30T1...
id (time) <U66 'S1A_IW_GRDH_1SDV_2022...
band <U2 'vh'
* x (x) float64 6.039e+05 ... 6.186e+05
* y (y) float64 1.624e+04 ... -1.095e+03
sar:resolution_range int64 20
... ...
title <U41 'VH: vertical transmit, horiz...
raster:bands object {'nodata': -32768, 'data_ty...
epsg int64 32647
gsd int64 30
proj:shape object {3600}
spatial_ref int64 0
Data variables:
vh (time, y, x) float16 dask.array<chunksize=(1, 579, 491), meta=np.ndarray>
vv (time, y, x) float16 dask.array<chunksize=(1, 579, 491), meta=np.ndarray>
dem (y, x) float16 dask.array<chunksize=(579, 491), meta=np.ndarray>
mask (y, x) uint8 0 0 0 0 0 ... 0 0 0 0 0dataslice = datacube.sel(time="2022-02-23T11:41:54.329096000").compute()
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(11, 12), sharex=True, sharey=True)
dataslice.vh.plot.imshow(ax=ax[0][0], cmap="bone", robust=True)
ax[0][0].set_title("Sentinel-1 RTC 20220223 VH")
dataslice.vv.plot.imshow(ax=ax[0][1], cmap="bone", robust=True)
ax[0][1].set_title("Sentinel-1 RTC 20220223 VV")
dataslice.dem.plot.imshow(ax=ax[1][0], cmap="gist_earth")
ax[1][0].set_title("Copernicus DEM")
dataslice.mask.plot.imshow(ax=ax[1][1], cmap="binary_r")
ax[1][1].set_title("Landslide mask")
plt.show()
2️⃣ Splitters and lumpers 🪨#
There are many ways to do change detection 🕵️. Here is but one ☝️.
Slice spatially and temporally 💇#
For the splitters, let’s first slice the datacube along the spatial dimension
into 256x256 chips 🍪 using zen3geo.datapipes.XbatcherSlicer
(functional name: slice_with_xbatcher). Refer to Chipping and batching data if you
need a 🧑🎓 refresher.
dp_xbatcher = dp_datacube.slice_with_xbatcher(input_dims={"y": 256, "x": 256})
dp_xbatcher
XbatcherSlicerIterDataPipe
Next, we’ll use the earthquake ⚠️ date to divide each 256x256 SAR time-series chip 🍕 with dimensions (time, y, x) into pre-event and post-event tensors. The target landslide 🛝 mask will be split out too.
def pre_post_target_tuple(
datachip: xr.Dataset, event_time: str = "2022-02-25T01:39:27"
) -> tuple[xr.Dataset, xr.Dataset, xr.Dataset]:
"""
From a single xarray.Dataset, split it into a tuple containing the
pre/post/target tensors.
"""
pre_times = datachip.time <= np.datetime64(event_time)
post_times = datachip.time > np.datetime64(event_time)
return (
datachip.sel(time=pre_times)[["vv", "vh", "dem"]],
datachip.sel(time=post_times)[["vv", "vh", "dem"]],
datachip[["mask"]],
)
dp_pre_post_target = dp_xbatcher.map(fn=pre_post_target_tuple)
dp_pre_post_target
MapperIterDataPipe
Inspect 👀 the shapes of one of the data chips that has been split into
pre/post/target 🍡 xarray.Dataset objects.
it = iter(dp_pre_post_target)
pre, post, target = next(it)
print(f"Before: {pre.sizes}")
print(f"After: {post.sizes}")
print(f"Target: {target.sizes}")
/home/docs/checkouts/readthedocs.org/user_builds/zen3geo/envs/v0.5.0/lib/python3.10/site-packages/torch/utils/data/datapipes/iter/combining.py:248: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
Before: Frozen({'time': 9, 'y': 256, 'x': 256})
After: Frozen({'time': 6, 'y': 256, 'x': 256})
Target: Frozen({'y': 256, 'x': 256})
Cool, at this point, you may want to decide 🤔 on how to handle different sized
before and after time-series images 🎞️. Or maybe not, and
torch.Tensor objects are all you desire ❤️🔥.
def dataset_to_tensors(triple_tuple) -> (torch.Tensor, torch.Tensor, torch.Tensor):
"""
Converts xarray.Datasets in a tuple into torch.Tensor objects.
"""
pre, post, target = triple_tuple
_pre: torch.Tensor = torch.as_tensor(pre.to_array().data)
_post: torch.Tensor = torch.as_tensor(post.to_array().data)
_target: torch.Tensor = torch.as_tensor(target.mask.data.astype("uint8"))
return _pre, _post, _target
dp_tensors = dp_pre_post_target.map(fn=dataset_to_tensors)
dp_tensors
MapperIterDataPipe
This is the final DataPipe graph ⛓️.
torchdata.datapipes.utils.to_graph(dp=dp_tensors)
Into a DataLoader 🏋️#
Time to connect the DataPipe to torch.utils.data.DataLoader ♻️!
dataloader = torch.utils.data.DataLoader2(dataset=dp_tensors, batch_size=None)
for i, batch in enumerate(dataloader):
pre, post, target = batch
print(f"Batch {i} - pre: {pre.shape}, post: {post.shape}, target: {target.shape}")
/home/docs/checkouts/readthedocs.org/user_builds/zen3geo/envs/v0.5.0/lib/python3.10/site-packages/torch/utils/data/datapipes/iter/combining.py:248: UserWarning: Some child DataPipes are not exhausted when __iter__ is called. We are resetting the buffer and each child DataPipe will read from the start again.
warnings.warn("Some child DataPipes are not exhausted when __iter__ is called. We are resetting "
Batch 0 - pre: torch.Size([3, 9, 256, 256]), post: torch.Size([3, 6, 256, 256]), target: torch.Size([256, 256])
Batch 1 - pre: torch.Size([3, 9, 256, 256]), post: torch.Size([3, 6, 256, 256]), target: torch.Size([256, 256])
Don’t just see change, be the change 🪧!
See also
This data pipeline is adapted from (a subset of) some amazing 🧪 research done during the Frontier Development Lab 2022 - Self Supervised Learning on SAR data for Change Detection challenge 🚀. Watch the final showcase video at https://www.youtube.com/watch?v=igAUTJwbmsY 📽️!