Multi-resolution#
On top of a hundred foot pole you linger
Clinging to the first mark of the scale
How do you proceed higher?
It will take more than a leap of faith
Earth Observation 🛰️ and climate projection 🌡️ data can be captured at different levels of detail. In this lesson, we’ll work with a multitude of spatial resolutions 📏, learning to respect the ground sampling distance or native resolution 🔬 of the physical variable being measured, while 🪶 minimizing memory usage. By the end of the lesson, you should be able to:
Find 🔍 low and high spatial resolution climate datasets and load them from Zarr stores
Stack 🥞 and subset time-series datasets with different spatial resolutions stored in a hierarchical
datatree.DataTreestructureSlice 🔪 the multi-resolution dataset along the time-axis into monthly bins
🔗 Links:
🎉 Getting started#
These are the tools 🛠️ you’ll need.
import matplotlib.pyplot as plt
import pandas as pd
import torchdata.dataloader2
import xarray as xr
import xpystac
import zen3geo
from datatree import DataTree
0️⃣ Find climate model datasets 🪸#
The two datasets we’ll be working with are 🌐 gridded climate projections, one that is in its original low 🔅 spatial resolution, and another one of a higher 🔆 spatial resolution. Specifically, we’ll be looking at the maximum temperature 🌡️ (tasmax) variable from one of the Coupled Model Intercomparison Project Phase 6 (CMIP6) global coupled ocean-atmosphere general circulation model (GCM) 💨 outputs that is of low-resolution (67.5 arcminute), and a super-resolution product from DeepSD 🤔 that is of a higher resolution (15 arcminute).
Note
The following tutorial will mostly use the term super-resolution 🔭 from Computer Vision instead of downscaling ⏬. It’s just that the term downscaling ⏬ (going from low to high resolution) can get confused with downsampling 🙃 (going from high to low resolution), whereas super-resolution 🔭 is unambiguously about going from low 🔅 to high 🔆 resolution.
🔖 References:
lowres_raw = "https://cpdataeuwest.blob.core.windows.net/cp-cmip/cmip6/ScenarioMIP/MRI/MRI-ESM2-0/ssp585/r1i1p1f1/Amon/tasmax/gn/v20191108"
highres_deepsd = "https://cpdataeuwest.blob.core.windows.net/cp-cmip/version1/data/DeepSD/ScenarioMIP.MRI.MRI-ESM2-0.ssp585.r1i1p1f1.month.DeepSD.tasmax.zarr"
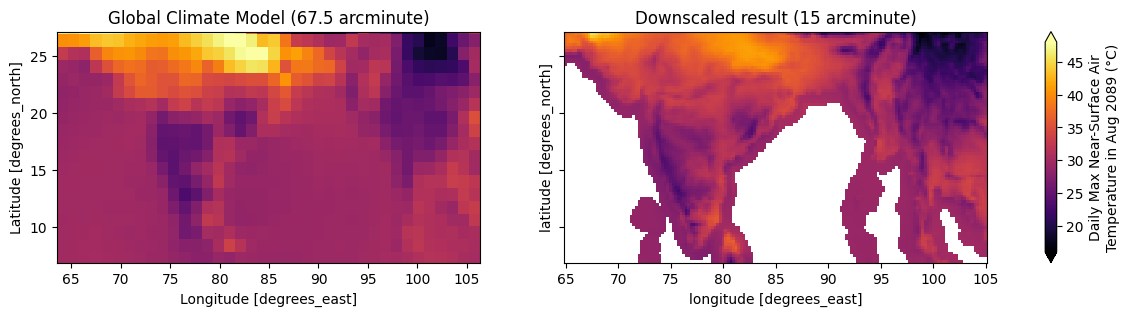
This is how the projected maximum temperature 🥵 for August 2089 looks like over South Asia 🪷 for a low-resolution 🔅 Global Climate Model (left) and a high-resolution 🔆 downscaled product (right).
Show code cell source
# Zarr datasets from https://github.com/carbonplan/research/blob/d05d148fd716ba6304e3833d765069dd890eaf4a/articles/cmip6-downscaling-explainer/components/downscaled-data.js#L97-L122
ds_gcm = xr.open_dataset(
filename_or_obj="https://cmip6downscaling.blob.core.windows.net/vis/article/fig1/regions/india/gcm-tasmax.zarr"
)
ds_gcm -= 273.15 # convert from Kelvin to Celsius
ds_downscaled = xr.open_dataset(
filename_or_obj="https://cmip6downscaling.blob.core.windows.net/vis/article/fig1/regions/india/downscaled-tasmax.zarr"
)
ds_downscaled -= 273.15 # convert from Kelvin to Celsius
# Plot projected maximum temperature over South Asia from GCM and GARD-MV
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(15, 3), sharey=True)
img1 = ds_gcm.tasmax.plot.imshow(
ax=ax[0], cmap="inferno", vmin=16, vmax=48, add_colorbar=False
)
ax[0].set_title("Global Climate Model (67.5 arcminute)")
img2 = ds_downscaled.tasmax.plot.imshow(
ax=ax[1], cmap="inferno", vmin=16, vmax=48, add_colorbar=False
)
ax[1].set_title("Downscaled result (15 arcminute)")
cbar = fig.colorbar(mappable=img1, ax=ax.ravel().tolist(), extend="both")
cbar.set_label(label="Daily Max Near-Surface Air\nTemperature in Aug 2089 (°C)")
plt.show()
Load Zarr stores 📦#
The Zarr stores 🧊 can be loaded into an
xarray.Dataset via zen3geo.datapipes.XpySTACAssetReader
(functional name: read_from_xpystac) with the engine="zarr" keyword
argument.
dp_lowres = torchdata.datapipes.iter.IterableWrapper(iterable=[lowres_raw])
dp_highres = torchdata.datapipes.iter.IterableWrapper(iterable=[highres_deepsd])
dp_lowres_dataset = dp_lowres.read_from_xpystac(engine="zarr", chunks="auto")
dp_highres_dataset = dp_highres.read_from_xpystac(engine="zarr", chunks="auto")
Inspect the climate datasets 🔥#
Let’s now preview 👀 the low-resolution 🔅 and high-resolution 🔆 temperature datasets.
it = iter(dp_lowres_dataset)
ds_lowres = next(it)
ds_lowres
<xarray.Dataset> Size: 211MB
Dimensions: (lat: 160, bnds: 2, lon: 320, time: 1032)
Coordinates:
height float64 8B ...
* lat (lat) float64 1kB -89.14 -88.03 -86.91 ... 86.91 88.03 89.14
lat_bnds (lat, bnds) float64 3kB dask.array<chunksize=(160, 2), meta=np.ndarray>
* lon (lon) float64 3kB 0.0 1.125 2.25 3.375 ... 356.6 357.8 358.9
lon_bnds (lon, bnds) float64 5kB dask.array<chunksize=(320, 2), meta=np.ndarray>
* time (time) datetime64[ns] 8kB 2015-01-16T12:00:00 ... 2100-12-16T1...
time_bnds (time, bnds) datetime64[ns] 17kB dask.array<chunksize=(1032, 2), meta=np.ndarray>
Dimensions without coordinates: bnds
Data variables:
tasmax (time, lat, lon) float32 211MB dask.array<chunksize=(516, 160, 320), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7 CMIP-6.2
activity_id: ScenarioMIP
branch_method: standard
branch_time_in_child: 60265.0
branch_time_in_parent: 60265.0
cmor_version: 3.4.0
... ...
table_info: Creation Date:(14 December 2018) MD5:b2d32d1a0d9b...
title: MRI-ESM2-0 output prepared for CMIP6
tracking_id: hdl:21.14100/421f03b2-8cb7-4473-9d03-0f772c8969c4
variable_id: tasmax
variant_label: r1i1p1f1
version_id: v20191108it = iter(dp_highres_dataset)
ds_highres = next(it)
ds_highres
<xarray.Dataset> Size: 4GB
Dimensions: (lat: 720, lon: 1440, time: 1020)
Coordinates:
* lat (lat) float64 6kB -89.88 -89.62 -89.38 -89.12 ... 89.38 89.62 89.88
* lon (lon) float64 12kB -179.9 -179.6 -179.4 ... 179.4 179.6 179.9
* time (time) datetime64[ns] 8kB 2015-01-01 2015-02-01 ... 2099-12-01
Data variables:
tasmax (time, lat, lon) float32 4GB dask.array<chunksize=(1020, 144, 144), meta=np.ndarray>
Attributes: (12/17)
Conventions: CF-1.8
activity_id: ScenarioMIP
cmip6_downscaling_contact: hello@carbonplan.org
cmip6_downscaling_explainer: https://carbonplan.org/research/cmip6-do...
cmip6_downscaling_institution: CarbonPlan
cmip6_downscaling_license: CC-BY-4.0
... ...
institution_id: MRI
member_id: r1i1p1f1
references: Eyring, V., Bony, S., Meehl, G. A., Seni...
source_id: MRI-ESM2-0
timescale: day
variable_id: tasmaxNotice that the low-resolution 🔅 dataset has lon/lat pixels of shape (320, 160), whereas the high-resolution 🔆 dataset is of shape (1440, 720). So there has been a 4.5x increase 📈 in spatial resolution going from the raw GCM 🌐 grid to the super-resolution 🔭 DeepSD grid.
Shift from 0-360 to -180-180 🌐#
A sharp eye 👁️ would have noticed that the longitudinal range of the
low-resolution 🔅 and high-resolution 🔆 dataset are offset ↔️ by 180°, going
from 0° to 360° and -180° to +180° respectively. Let’s shift the coordinates 📍
of the low-resolution grid 🌍 from 0-360 to -180-180 using a custom
torchdata.datapipes.iter.Mapper (functional name: map) function.
🔖 References:
def shift_longitude_360_to_180(ds: xr.Dataset) -> xr.Dataset:
ds = ds.assign_coords(lon=(((ds.lon + 180) % 360) - 180))
ds = ds.roll(lon=int(len(ds.lon) / 2), roll_coords=True)
return ds
dp_lowres_dataset_180 = dp_lowres_dataset.map(fn=shift_longitude_360_to_180)
dp_lowres_dataset_180
MapperIterDataPipe
Double check that the low-resolution 🔆 grid’s longitude coordinates 🔢 are now in the -180° to +180° range.
it = iter(dp_lowres_dataset_180)
ds_lowres_180 = next(it)
ds_lowres_180
<xarray.Dataset> Size: 211MB
Dimensions: (lat: 160, bnds: 2, lon: 320, time: 1032)
Coordinates:
height float64 8B ...
* lat (lat) float64 1kB -89.14 -88.03 -86.91 ... 86.91 88.03 89.14
lat_bnds (lat, bnds) float64 3kB dask.array<chunksize=(160, 2), meta=np.ndarray>
lon_bnds (lon, bnds) float64 5kB dask.array<chunksize=(320, 2), meta=np.ndarray>
* time (time) datetime64[ns] 8kB 2015-01-16T12:00:00 ... 2100-12-16T1...
time_bnds (time, bnds) datetime64[ns] 17kB dask.array<chunksize=(1032, 2), meta=np.ndarray>
* lon (lon) float64 3kB -180.0 -178.9 -177.8 ... 176.6 177.8 178.9
Dimensions without coordinates: bnds
Data variables:
tasmax (time, lat, lon) float32 211MB dask.array<chunksize=(516, 160, 320), meta=np.ndarray>
Attributes: (12/47)
Conventions: CF-1.7 CMIP-6.2
activity_id: ScenarioMIP
branch_method: standard
branch_time_in_child: 60265.0
branch_time_in_parent: 60265.0
cmor_version: 3.4.0
... ...
table_info: Creation Date:(14 December 2018) MD5:b2d32d1a0d9b...
title: MRI-ESM2-0 output prepared for CMIP6
tracking_id: hdl:21.14100/421f03b2-8cb7-4473-9d03-0f772c8969c4
variable_id: tasmax
variant_label: r1i1p1f1
version_id: v20191108Spatiotemporal stack and subset 🍱#
Following on from Stacking layers where multiple 🥞 layers with the same
spatial resolution were stacked together into an xarray.DataArray
object, this section will teach 🧑🏫 you about stacking datasets with
different spatial resolutions 📶 into a datatree.DataTree
object that has a nested/hierarchical structure. That
datatree.DataTree can then be subsetted 🥮 to the desired spatial
and temporal extent in one go 😎.
Stack multi-resolution datasets 📚#
First, we’ll need to combine 🪢 the low-resolution GCM and high-resolution
DeepSD xarray.Dataset objects into a tuple 🎵 using
torchdata.datapipes.iter.Zipper (functional name: zip).
dp_lowres_highres = dp_lowres_dataset_180.zip(dp_highres_dataset)
dp_lowres_highres
ZipperIterDataPipe
Next, use torchdata.datapipes.iter.Collator (functional name:
collate) to convert 🤸 the tuple of xarray.Dataset objects into
an datatree.DataTree 🎋, similar to what was done in
Stacking layers. Note that we’ll only take the ‘tasmax’ ♨️ (Daily Maximum
Near-Surface Air Temperature) xarray.DataArray variable from each
of the xarray.Dataset objects.
def multires_collate_fn(lowres_and_highres: tuple) -> DataTree:
"""
Combine a pair of xarray.Dataset (lowres, highres) inputs into a
datatree.DataTree with groups named 'lowres' and 'highres'.
"""
# Turn 2 xr.Dataset objects into 1 xr.DataTree with multiple groups
ds_lowres, ds_highres = lowres_and_highres
# Create DataTree with lowres and highres groups
datatree: DataTree = DataTree.from_dict(
d={"lowres": ds_lowres.tasmax, "highres": ds_highres.tasmax}
)
return datatree
dp_datatree = dp_lowres_highres.collate(collate_fn=multires_collate_fn)
dp_datatree
CollatorIterDataPipe
See the nested 🪆 structure of the datatree.DataTree. The
low-resolution 🔅 GCM and high-resolution 🔆 DeepSD outputs have been placed in
separate groups 🖖.
it = iter(dp_datatree)
datatree = next(it)
datatree
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*Subset multi-resolution layers 🥮#
The climate model outputs above are a global 🗺️ one covering a timespan from
January 2015 to December 2100 📅. If you’re only interested in a particular
region 🌏 or timespan ⌚, then the datatree.DataTree will need to
be trimmed 💇 down. Let’s use datatree.DataTree.sel() to subset the
multi-resolution data to just the Philippines 🇵🇭 for the period 2015 to 2030.
def spatiotemporal_subset(dt: DataTree) -> DataTree:
dt_subset = dt.sel(
lon=slice(116.4375, 126.5625),
lat=slice(5.607445, 19.065325),
time=slice("2015-01-01", "2030-12-31"),
)
return dt_subset
dp_datatree_subset = dp_datatree.map(fn=spatiotemporal_subset)
dp_datatree_subset
MapperIterDataPipe
Inspect the subsetted climate dataset 🕵️
it = iter(dp_datatree_subset)
datatree_subset = next(it)
datatree_subset
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
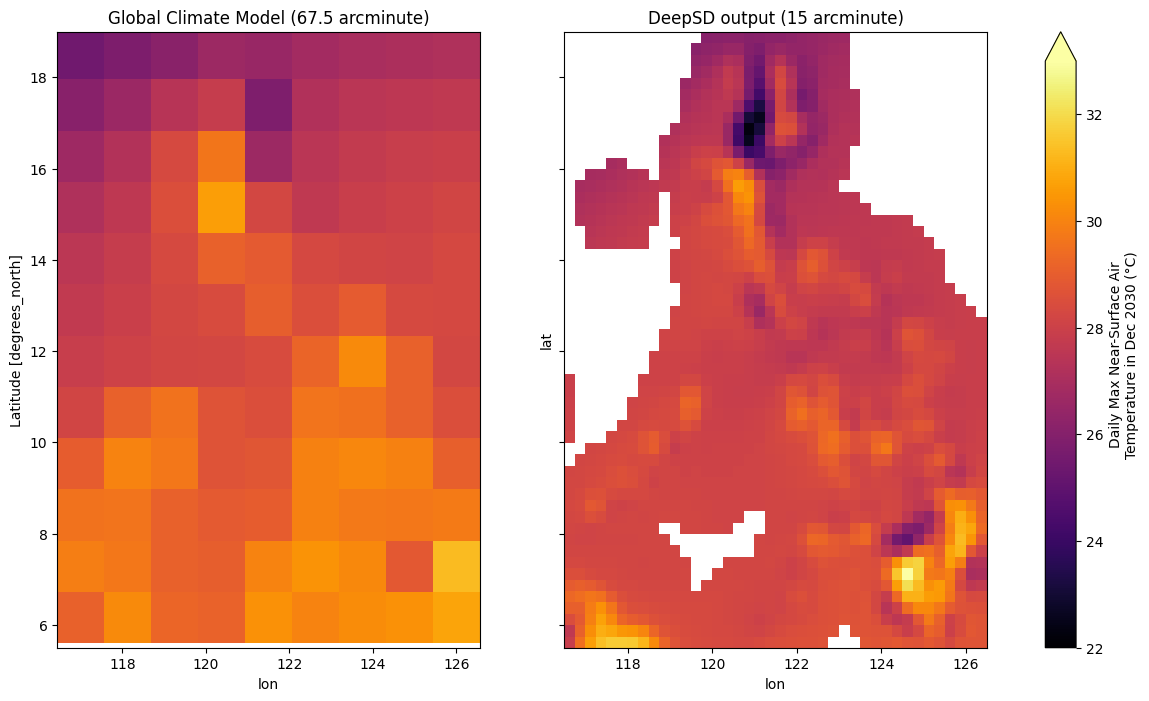
*empty*Let’s plot the projected temperature 🌡️ for Dec 2030 over the Philippine Archipelago to ensure things look ok.
ds_lowres = (
datatree_subset["lowres/tasmax"]
.sel(time=slice("2030-12-01", "2030-12-31"))
.squeeze()
)
ds_lowres -= 273.15 # convert from Kelvin to Celsius
ds_highres = (
datatree_subset["highres/tasmax"]
.sel(time=slice("2030-12-01", "2030-12-31"))
.squeeze()
)
ds_highres -= 273.15 # convert from Kelvin to Celsius
# Plot projected maximum temperature over the Philippines from GCM and DeepSD
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(15, 8), sharey=True)
img1 = ds_lowres.plot.imshow(
ax=ax[0], cmap="inferno", vmin=22, vmax=33, add_colorbar=False
)
ax[0].set_title("Global Climate Model (67.5 arcminute)")
img2 = ds_highres.plot.imshow(
ax=ax[1], cmap="inferno", vmin=22, vmax=33, add_colorbar=False
)
ax[1].set_title("DeepSD output (15 arcminute)")
cbar = fig.colorbar(mappable=img1, ax=ax.ravel().tolist(), extend="max")
cbar.set_label(label="Daily Max Near-Surface Air\nTemperature in Dec 2030 (°C)")
plt.show()
Important
When slicing ✂️ different spatial resolution grids, put some 🧠 thought into the process. Do some 🧮 math to ensure the coordinates of the bounding box (min/max lon/lat) cut through the pixels exactly at the 📐 pixel boundaries whenever possible.
If your multi-resolution 📶 layers have spatial resolutions that are round multiples ✖️ of each other (e.g. 10m, 20m, 60m), it is advisable to align 🎯 the pixel corners, such that the high-resolution 🔆 pixels fit within the low-resolution 🔅 pixels (e.g. one 20m pixel should contain four 10m pixels). This can be done by resampling 🖌️ or interpolating the grid (typically the higher resolution one) onto a new reference frame 🖼️.
For datasets ℹ️ that come from different sources and need to be reprojected 🔁, you can do the reprojection and pixel alignment in a single step 🔂. Be extra careful about resampling, as certain datasets (e.g. complex SAR 📡 data that has been collected off-nadir) may require special 🌷 treatment.
Time to slice again ⌛#
So, we now have a datatree.DataTree with two 💕 groups/nodes called
‘lowres’ and ‘highres’ that have tensor shapes (lat: 12, lon: 9, time: 192)
and (lat: 54, lon: 40, time: 192) respectively. While the time dimension ⏱️
is of the same length, the timestamp values between the low-resolution 🔅 GCM
and high-resolution 🔆 DeepSD output are different. Specifically, the GCM
output dates at the middle of the month 📅, while the DeepSD output has dates
at the start of the month. Let’s see how this can be handled 🫖.
Slicing by month 🗓️#
Assuming that the roughly two week offset ↔️ between the monthly resolution GCM
and DeepSD time-series is negligible 🤏, we can split the dataset on the time
dimension at the start/end of each month 📆. Let’s write a function and use
torchdata.datapipes.iter.FlatMapper (functional name: flatmap)
for this.
def split_on_month(dt: DataTree, node:str = "highres/tasmax") -> DataTree:
"""
Return a slice of data for every month in a datatree.DataTree time-series.
"""
for t in dt[node].time.to_pandas():
dt_slice = dt.sel(
time=slice(t + pd.offsets.MonthBegin(0), t + pd.offsets.MonthEnd(0))
)
yield dt_slice.squeeze(dim="time")
dp_datatree_timeslices = dp_datatree_subset.flatmap(fn=split_on_month)
dp_datatree_timeslices
FlatMapperIterDataPipe
The datapipe should yield a datatree.DataTree with just one
month’s 📅 worth of temperature 🌡️ data per iteration.
it = iter(dp_datatree_timeslices)
datatree_timeslice = next(it)
datatree_timeslice
<xarray.DatasetView> Size: 0B
Dimensions: ()
Data variables:
*empty*See also
Those interested in slicing multi-resolution arrays spatially can keep an eye
on the 🚧 ongoing implementation at
xarray-contrib/xbatcher#171 and the discussion at
xarray-contrib/xbatcher#93. This 🧑🏫 tutorial will be
updated ♻️ once there’s a clean way to generate multi-resolution
datatree.DataTree slices in a newer release of
xbatcher 😉
Visualize the final DataPipe graph ⛓️.
torchdata.datapipes.utils.to_graph(dp=dp_datatree_timeslices)
Into a DataLoader 🏋️#
Ready to populate the torchdata.dataloader2.DataLoader2 🏭!
dataloader = torchdata.dataloader2.DataLoader2(datapipe=dp_datatree_timeslices)
for i, batch in enumerate(dataloader):
ds_lowres = batch["lowres/tasmax"]
ds_highres = batch["highres/tasmax"]
print(f"Batch {i} - lowres: {ds_lowres.shape}, highres: {ds_highres.shape}")
if i > 8:
break
Batch 0 - lowres: (12, 9), highres: (54, 40)
Batch 1 - lowres: (12, 9), highres: (54, 40)
Batch 2 - lowres: (12, 9), highres: (54, 40)
Batch 3 - lowres: (12, 9), highres: (54, 40)
Batch 4 - lowres: (12, 9), highres: (54, 40)
Batch 5 - lowres: (12, 9), highres: (54, 40)
Batch 6 - lowres: (12, 9), highres: (54, 40)
Batch 7 - lowres: (12, 9), highres: (54, 40)
Batch 8 - lowres: (12, 9), highres: (54, 40)
Batch 9 - lowres: (12, 9), highres: (54, 40)
Do super-resolution, but make no illusion 🧚
See also
Credits to CarbonPlan for making the code and data for their CMIP6 downscaling work openly available. Find out more at https://docs.carbonplan.org/cmip6-downscaling!